Reference-based Enterotype Classification Tool
Enterotypes describe clustering patterns across samples from the human intestinal microbiome that are associated with disease, medication, diet, and lifestyle. The Enterotyper is a computational tool designed for the scientific community to classify human fecal metagenomes into established enterotypes. Using a robust XGboost machine learning model, this tool provides accurate enterotype classification based on a large global dataset of fecal metagenomes.

Please cite when you use Enterotyper: Keller et al.
Usage
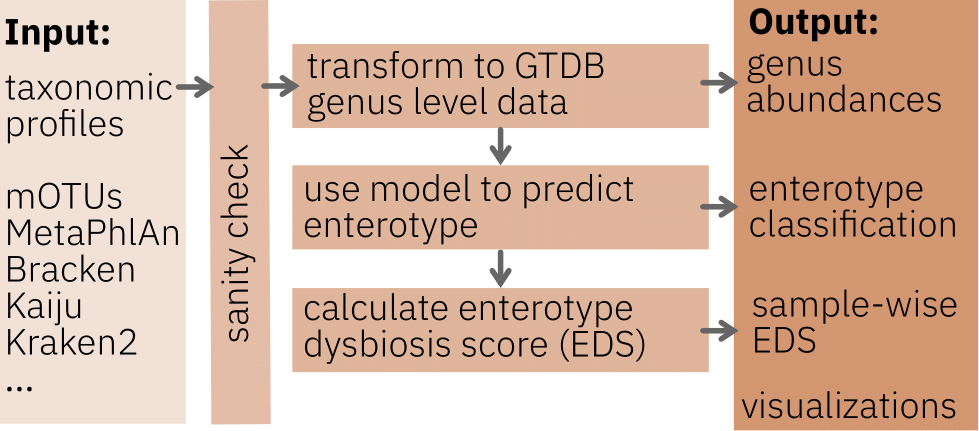
Input
Please upload a taxonomic profile generated either by one of the following taxonomic profiler:
Alternatively, you can also upload a GTDB taxonomic profile at genus level with genera as rownames and sample names as columns. An example file is shown here.
Output
EnterotypeAssignment_EnterotypeModel.tsvcontains the enterotype classification. In case of FKM clustering, a classification strength for each enterotype for each sample is also reported. The enterotype with the highest classification strength determines the hard classification. In addition, the Enterotype Dysbiosis Score, which is the inverted, z-score normalised and scaled between 0 and 1 maximum classification strength, is reported in the FKM 3-enterotype model.GTDB_genus_level_taxonomy.tsvcontains the GTD genus-level taxonomy profiles for the uploaded data which is used as a basis for the Enterotyper.- Visualizations displayed on the web page for a first assessment of the predicted enterotype classification of the input data and comparison with the enterotype landscape in the global dataset used to build the Enterotyper.
Troubleshooting
Try the following if you encounter errors:- Make sure the taxonomic profiler name and version are in the first line of the file after a hash (#). e.g.:
# motus version 3.0.1 - If you're supplying a genus level file, please ensure the following:
- The line with sample names must NOT be commented out with a hash (#) e.g.
`# g__genus sample1 sample2`would be incorrect - The line with sample names must start with
g__genus - Samples names must NOT have an underscore ( _ ) in the name